ultralytics 8.0.105 classification hyp fix and new onplot callbacks (#2684)
Co-authored-by: ayush chaurasia <ayush.chaurarsia@gmail.com> Co-authored-by: pre-commit-ci[bot] <66853113+pre-commit-ci[bot]@users.noreply.github.com> Co-authored-by: Ivan Shcheklein <shcheklein@gmail.com>
This commit is contained in:
@ -33,9 +33,18 @@ def extract_classes_and_functions(filepath):
|
|||||||
def create_markdown(py_filepath, module_path, classes, functions):
|
def create_markdown(py_filepath, module_path, classes, functions):
|
||||||
md_filepath = py_filepath.with_suffix('.md')
|
md_filepath = py_filepath.with_suffix('.md')
|
||||||
|
|
||||||
|
# Read existing content and keep header content between first two ---
|
||||||
|
header_content = ""
|
||||||
|
if md_filepath.exists():
|
||||||
|
with open(md_filepath, 'r') as file:
|
||||||
|
existing_content = file.read()
|
||||||
|
header_parts = existing_content.split('---', 2)
|
||||||
|
if len(header_parts) >= 3:
|
||||||
|
header_content = f"{header_parts[0]}---{header_parts[1]}---\n\n"
|
||||||
|
|
||||||
md_content = [f"# {class_name}\n---\n:::{module_path}.{class_name}\n<br><br>\n" for class_name in classes]
|
md_content = [f"# {class_name}\n---\n:::{module_path}.{class_name}\n<br><br>\n" for class_name in classes]
|
||||||
md_content.extend(f"# {func_name}\n---\n:::{module_path}.{func_name}\n<br><br>\n" for func_name in functions)
|
md_content.extend(f"# {func_name}\n---\n:::{module_path}.{func_name}\n<br><br>\n" for func_name in functions)
|
||||||
md_content = "\n".join(md_content)
|
md_content = header_content + "\n".join(md_content)
|
||||||
|
|
||||||
os.makedirs(os.path.dirname(md_filepath), exist_ok=True)
|
os.makedirs(os.path.dirname(md_filepath), exist_ok=True)
|
||||||
with open(md_filepath, 'w') as file:
|
with open(md_filepath, 'w') as file:
|
||||||
|
|||||||
@ -1,7 +1,81 @@
|
|||||||
---
|
---
|
||||||
comments: true
|
comments: true
|
||||||
|
description: Learn about the MNIST dataset, a large database of handwritten digits commonly used for training various image processing systems and machine learning models.
|
||||||
---
|
---
|
||||||
|
|
||||||
# 🚧 Page Under Construction ⚒
|
# MNIST Dataset
|
||||||
|
|
||||||
This page is currently under construction!️ 👷Please check back later for updates. 😃🔜
|
The [MNIST](http://yann.lecun.com/exdb/mnist/) (Modified National Institute of Standards and Technology) dataset is a large database of handwritten digits that is commonly used for training various image processing systems and machine learning models. It was created by "re-mixing" the samples from NIST's original datasets and has become a benchmark for evaluating the performance of image classification algorithms.
|
||||||
|
|
||||||
|
## Key Features
|
||||||
|
|
||||||
|
- MNIST contains 60,000 training images and 10,000 testing images of handwritten digits.
|
||||||
|
- The dataset comprises grayscale images of size 28x28 pixels.
|
||||||
|
- The images are normalized to fit into a 28x28 pixel bounding box and anti-aliased, introducing grayscale levels.
|
||||||
|
- MNIST is widely used for training and testing in the field of machine learning, especially for image classification tasks.
|
||||||
|
|
||||||
|
## Dataset Structure
|
||||||
|
|
||||||
|
The MNIST dataset is split into two subsets:
|
||||||
|
|
||||||
|
1. **Training Set**: This subset contains 60,000 images of handwritten digits used for training machine learning models.
|
||||||
|
2. **Testing Set**: This subset consists of 10,000 images used for testing and benchmarking the trained models.
|
||||||
|
|
||||||
|
## Extended MNIST (EMNIST)
|
||||||
|
|
||||||
|
Extended MNIST (EMNIST) is a newer dataset developed and released by NIST to be the successor to MNIST. While MNIST included images only of handwritten digits, EMNIST includes all the images from NIST Special Database 19, which is a large database of handwritten uppercase and lowercase letters as well as digits. The images in EMNIST were converted into the same 28x28 pixel format, by the same process, as were the MNIST images. Accordingly, tools that work with the older, smaller MNIST dataset will likely work unmodified with EMNIST.
|
||||||
|
|
||||||
|
## Applications
|
||||||
|
|
||||||
|
The MNIST dataset is widely used for training and evaluating deep learning models in image classification tasks, such as Convolutional Neural Networks (CNNs), Support Vector Machines (SVMs), and various other machine learning algorithms. The dataset's simple and well-structured format makes it an essential resource for researchers and practitioners in the field of machine learning and computer vision.
|
||||||
|
|
||||||
|
## Usage
|
||||||
|
|
||||||
|
To train a CNN model on the MNIST dataset for 100 epochs with an image size of 32x32, you can use the following code snippets. For a comprehensive list of available arguments, refer to the model [Training](../../modes/train.md) page.
|
||||||
|
|
||||||
|
!!! example "Train Example"
|
||||||
|
|
||||||
|
=== "Python"
|
||||||
|
|
||||||
|
```python
|
||||||
|
from ultralytics import YOLO
|
||||||
|
|
||||||
|
# Load a model
|
||||||
|
model = YOLO('yolov8n-cls.pt') # load a pretrained model (recommended for training)
|
||||||
|
|
||||||
|
# Train the model
|
||||||
|
model.train(data='mnist', epochs=100, imgsz=32)
|
||||||
|
```
|
||||||
|
|
||||||
|
=== "CLI"
|
||||||
|
|
||||||
|
```bash
|
||||||
|
# Start training from a pretrained *.pt model
|
||||||
|
cnn detect train data=MNIST.yaml model=cnn_mnist.pt epochs=100 imgsz=28
|
||||||
|
```
|
||||||
|
|
||||||
|
## Sample Images and Annotations
|
||||||
|
|
||||||
|
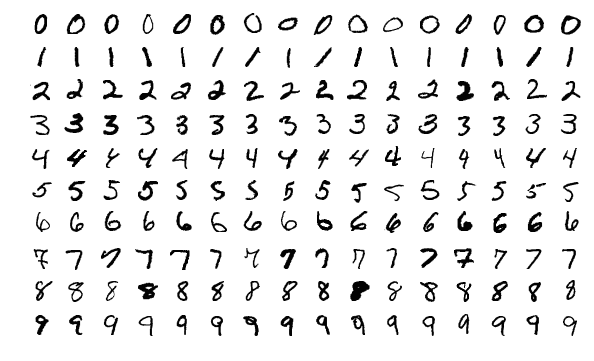
The MNIST dataset contains grayscale images of handwritten digits, providing a well-structured dataset for image classification tasks. Here are some examples of images from the dataset:
|
||||||
|
|
||||||
|
|
||||||
|
|
||||||
|
The example showcases the variety and complexity of the handwritten digits in the MNIST dataset, highlighting the importance of a diverse dataset for training robust image classification models.
|
||||||
|
|
||||||
|
## Citations and Acknowledgments
|
||||||
|
|
||||||
|
If you use the MNIST dataset in your
|
||||||
|
|
||||||
|
research or development work, please cite the following paper:
|
||||||
|
|
||||||
|
```bibtex
|
||||||
|
@article{lecun2010mnist,
|
||||||
|
title={MNIST handwritten digit database},
|
||||||
|
author={LeCun, Yann and Cortes, Corinna and Burges, CJ},
|
||||||
|
journal={ATT Labs [Online]. Available: http://yann.lecun.com/exdb/mnist},
|
||||||
|
volume={2},
|
||||||
|
year={2010}
|
||||||
|
}
|
||||||
|
```
|
||||||
|
|
||||||
|
We would like to acknowledge Yann LeCun, Corinna Cortes, and Christopher J.C. Burges for creating and maintaining the MNIST dataset as a valuable resource for the machine learning and computer vision research community. For more information about the MNIST dataset and its creators, visit the [MNIST dataset website](http://yann.lecun.com/exdb/mnist/).
|
||||||
@ -118,7 +118,7 @@ Auto-annotation is an essential feature that allows you to generate a segmentati
|
|||||||
To auto-annotate your dataset using the Ultralytics framework, you can use the `auto_annotate` function as shown below:
|
To auto-annotate your dataset using the Ultralytics framework, you can use the `auto_annotate` function as shown below:
|
||||||
|
|
||||||
```python
|
```python
|
||||||
from ultralytics.yolo.data import auto_annotate
|
from ultralytics.yolo.data.annotator import auto_annotate
|
||||||
|
|
||||||
auto_annotate(data="path/to/images", det_model="yolov8x.pt", sam_model='sam_b.pt')
|
auto_annotate(data="path/to/images", det_model="yolov8x.pt", sam_model='sam_b.pt')
|
||||||
```
|
```
|
||||||
|
|||||||
@ -9,6 +9,12 @@ description: Explore RT-DETR, a high-performance real-time object detector. Lear
|
|||||||
|
|
||||||
Real-Time Detection Transformer (RT-DETR) is an end-to-end object detector that provides real-time performance while maintaining high accuracy. It efficiently processes multi-scale features by decoupling intra-scale interaction and cross-scale fusion, and supports flexible adjustment of inference speed using different decoder layers without retraining. RT-DETR outperforms many real-time object detectors on accelerated backends like CUDA with TensorRT.
|
Real-Time Detection Transformer (RT-DETR) is an end-to-end object detector that provides real-time performance while maintaining high accuracy. It efficiently processes multi-scale features by decoupling intra-scale interaction and cross-scale fusion, and supports flexible adjustment of inference speed using different decoder layers without retraining. RT-DETR outperforms many real-time object detectors on accelerated backends like CUDA with TensorRT.
|
||||||
|
|
||||||
|
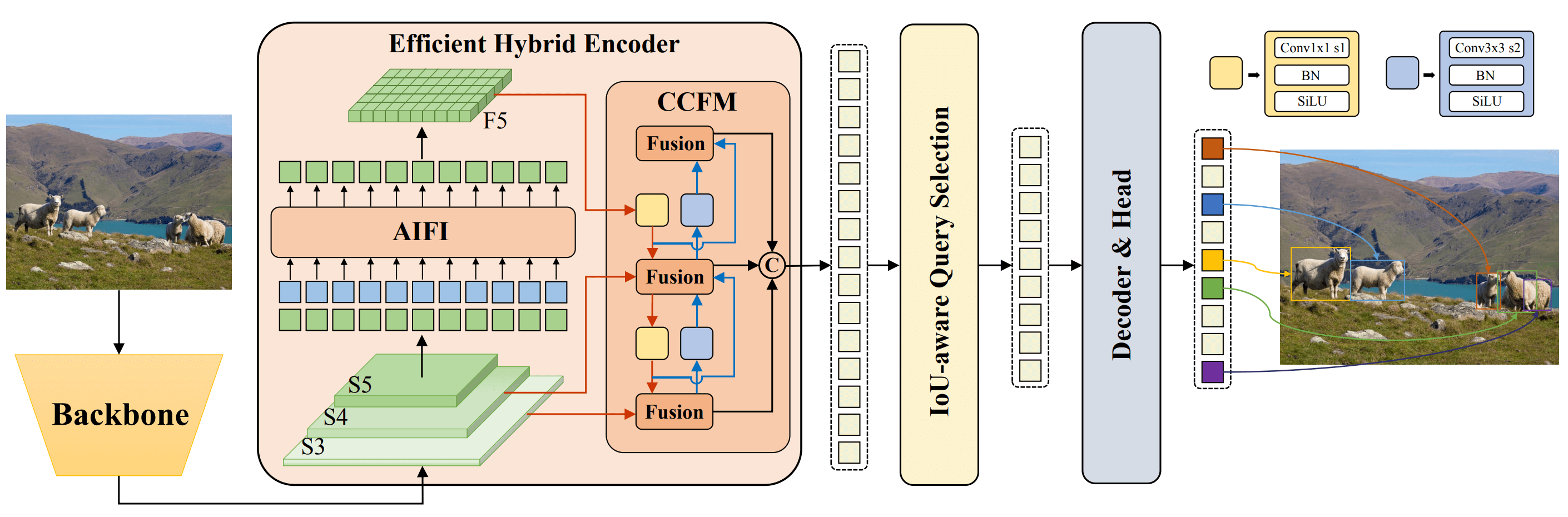
|
||||||
|
**Overview of RT-DETR.** Model architecture diagram showing the last three stages of the backbone {S3, S4, S5} as the input
|
||||||
|
to the encoder. The efficient hybrid encoder transforms multiscale features into a sequence of image features through intrascale feature interaction (AIFI) and cross-scale feature-fusion module (CCFM). The IoU-aware query selection is employed
|
||||||
|
to select a fixed number of image features to serve as initial object queries for the decoder. Finally, the decoder with auxiliary
|
||||||
|
prediction heads iteratively optimizes object queries to generate boxes and confidence scores ([source](https://arxiv.org/pdf/2304.08069.pdf)).
|
||||||
|
|
||||||
### Key Features
|
### Key Features
|
||||||
|
|
||||||
- **Efficient Hybrid Encoder:** RT-DETR uses an efficient hybrid encoder that processes multi-scale features by decoupling intra-scale interaction and cross-scale fusion. This design reduces computational costs and allows for real-time object detection.
|
- **Efficient Hybrid Encoder:** RT-DETR uses an efficient hybrid encoder that processes multi-scale features by decoupling intra-scale interaction and cross-scale fusion. This design reduces computational costs and allows for real-time object detection.
|
||||||
|
|||||||
@ -57,7 +57,7 @@ Auto-annotation is an essential feature that allows you to generate a [segmentat
|
|||||||
To auto-annotate your dataset using the Ultralytics framework, you can use the `auto_annotate` function as shown below:
|
To auto-annotate your dataset using the Ultralytics framework, you can use the `auto_annotate` function as shown below:
|
||||||
|
|
||||||
```python
|
```python
|
||||||
from ultralytics.yolo.data import auto_annotate
|
from ultralytics.yolo.data.annotator import auto_annotate
|
||||||
|
|
||||||
auto_annotate(data="path/to/images", det_model="yolov8x.pt", sam_model='sam_b.pt')
|
auto_annotate(data="path/to/images", det_model="yolov8x.pt", sam_model='sam_b.pt')
|
||||||
```
|
```
|
||||||
|
|||||||
19
tests/conftest.py
Normal file
19
tests/conftest.py
Normal file
@ -0,0 +1,19 @@
|
|||||||
|
import pytest
|
||||||
|
|
||||||
|
|
||||||
|
def pytest_addoption(parser):
|
||||||
|
parser.addoption('--runslow', action='store_true', default=False, help='run slow tests')
|
||||||
|
|
||||||
|
|
||||||
|
def pytest_configure(config):
|
||||||
|
config.addinivalue_line('markers', 'slow: mark test as slow to run')
|
||||||
|
|
||||||
|
|
||||||
|
def pytest_collection_modifyitems(config, items):
|
||||||
|
if config.getoption('--runslow'):
|
||||||
|
# --runslow given in cli: do not skip slow tests
|
||||||
|
return
|
||||||
|
skip_slow = pytest.mark.skip(reason='need --runslow option to run')
|
||||||
|
for item in items:
|
||||||
|
if 'slow' in item.keywords:
|
||||||
|
item.add_marker(skip_slow)
|
||||||
@ -3,10 +3,17 @@
|
|||||||
import subprocess
|
import subprocess
|
||||||
from pathlib import Path
|
from pathlib import Path
|
||||||
|
|
||||||
from ultralytics.yolo.utils import LINUX, ONLINE, ROOT, SETTINGS
|
import pytest
|
||||||
|
|
||||||
MODEL = Path(SETTINGS['weights_dir']) / 'yolov8n'
|
from ultralytics.yolo.utils import ONLINE, ROOT, SETTINGS
|
||||||
CFG = 'yolov8n'
|
|
||||||
|
WEIGHT_DIR = Path(SETTINGS['weights_dir'])
|
||||||
|
TASK_ARGS = [ # (task, model, data)
|
||||||
|
('detect', 'yolov8n', 'coco8.yaml'), ('segment', 'yolov8n-seg', 'coco8-seg.yaml'),
|
||||||
|
('classify', 'yolov8n-cls', 'imagenet10'), ('pose', 'yolov8n-pose', 'coco8-pose.yaml')]
|
||||||
|
EXPORT_ARGS = [ # (model, format)
|
||||||
|
('yolov8n', 'torchscript'), ('yolov8n-seg', 'torchscript'), ('yolov8n-cls', 'torchscript'),
|
||||||
|
('yolov8n-pose', 'torchscript')]
|
||||||
|
|
||||||
|
|
||||||
def run(cmd):
|
def run(cmd):
|
||||||
@ -20,78 +27,33 @@ def test_special_modes():
|
|||||||
run('yolo help')
|
run('yolo help')
|
||||||
|
|
||||||
|
|
||||||
# Train checks ---------------------------------------------------------------------------------------------------------
|
@pytest.mark.parametrize('task,model,data', TASK_ARGS)
|
||||||
def test_train_det():
|
def test_train(task, model, data):
|
||||||
run(f'yolo train detect model={CFG}.yaml data=coco8.yaml imgsz=32 epochs=1 v5loader')
|
run(f'yolo train {task} model={model}.yaml data={data} imgsz=32 epochs=1')
|
||||||
|
|
||||||
|
|
||||||
def test_train_seg():
|
@pytest.mark.parametrize('task,model,data', TASK_ARGS)
|
||||||
run(f'yolo train segment model={CFG}-seg.yaml data=coco8-seg.yaml imgsz=32 epochs=1')
|
def test_val(task, model, data):
|
||||||
|
run(f'yolo val {task} model={model}.pt data={data} imgsz=32')
|
||||||
|
|
||||||
|
|
||||||
def test_train_cls():
|
@pytest.mark.parametrize('task,model,data', TASK_ARGS)
|
||||||
run(f'yolo train classify model={CFG}-cls.yaml data=imagenet10 imgsz=32 epochs=1')
|
def test_predict(task, model, data):
|
||||||
|
run(f"yolo predict model={model}.pt source={ROOT / 'assets'} imgsz=32 save save_crop save_txt")
|
||||||
|
|
||||||
def test_train_pose():
|
|
||||||
run(f'yolo train pose model={CFG}-pose.yaml data=coco8-pose.yaml imgsz=32 epochs=1')
|
|
||||||
|
|
||||||
|
|
||||||
# Val checks -----------------------------------------------------------------------------------------------------------
|
|
||||||
def test_val_detect():
|
|
||||||
run(f'yolo val detect model={MODEL}.pt data=coco8.yaml imgsz=32')
|
|
||||||
|
|
||||||
|
|
||||||
def test_val_segment():
|
|
||||||
run(f'yolo val segment model={MODEL}-seg.pt data=coco8-seg.yaml imgsz=32')
|
|
||||||
|
|
||||||
|
|
||||||
def test_val_classify():
|
|
||||||
run(f'yolo val classify model={MODEL}-cls.pt data=imagenet10 imgsz=32')
|
|
||||||
|
|
||||||
|
|
||||||
def test_val_pose():
|
|
||||||
run(f'yolo val pose model={MODEL}-pose.pt data=coco8-pose.yaml imgsz=32')
|
|
||||||
|
|
||||||
|
|
||||||
# Predict checks -------------------------------------------------------------------------------------------------------
|
|
||||||
def test_predict_detect():
|
|
||||||
run(f"yolo predict model={MODEL}.pt source={ROOT / 'assets'} imgsz=32 save save_crop save_txt")
|
|
||||||
if ONLINE:
|
if ONLINE:
|
||||||
run(f'yolo predict model={MODEL}.pt source=https://ultralytics.com/images/bus.jpg imgsz=32')
|
run(f'yolo predict model={model}.pt source=https://ultralytics.com/images/bus.jpg imgsz=32')
|
||||||
run(f'yolo predict model={MODEL}.pt source=https://ultralytics.com/assets/decelera_landscape_min.mov imgsz=32')
|
run(f'yolo predict model={model}.pt source=https://ultralytics.com/assets/decelera_landscape_min.mov imgsz=32')
|
||||||
run(f'yolo predict model={MODEL}.pt source=https://ultralytics.com/assets/decelera_portrait_min.mov imgsz=32')
|
run(f'yolo predict model={model}.pt source=https://ultralytics.com/assets/decelera_portrait_min.mov imgsz=32')
|
||||||
|
|
||||||
|
|
||||||
def test_predict_segment():
|
@pytest.mark.parametrize('model,format', EXPORT_ARGS)
|
||||||
run(f"yolo predict model={MODEL}-seg.pt source={ROOT / 'assets'} imgsz=32 save save_txt")
|
def test_export(model, format):
|
||||||
|
run(f'yolo export model={model}.pt format={format}')
|
||||||
|
|
||||||
|
|
||||||
def test_predict_classify():
|
# Slow Tests
|
||||||
run(f"yolo predict model={MODEL}-cls.pt source={ROOT / 'assets'} imgsz=32 save save_txt")
|
@pytest.mark.slow
|
||||||
|
@pytest.mark.parametrize('task,model,data', TASK_ARGS)
|
||||||
|
def test_train_gpu(task, model, data):
|
||||||
def test_predict_pose():
|
run(f'yolo train {task} model={model}.yaml data={data} imgsz=32 epochs=1 device="0"') # single GPU
|
||||||
run(f"yolo predict model={MODEL}-pose.pt source={ROOT / 'assets'} imgsz=32 save save_txt")
|
run(f'yolo train {task} model={model}.pt data={data} imgsz=32 epochs=1 device="0,1"') # Multi GPU
|
||||||
|
|
||||||
|
|
||||||
# Export checks --------------------------------------------------------------------------------------------------------
|
|
||||||
def test_export_detect_torchscript():
|
|
||||||
run(f'yolo export model={MODEL}.pt format=torchscript')
|
|
||||||
|
|
||||||
|
|
||||||
def test_export_segment_torchscript():
|
|
||||||
run(f'yolo export model={MODEL}-seg.pt format=torchscript')
|
|
||||||
|
|
||||||
|
|
||||||
def test_export_classify_torchscript():
|
|
||||||
run(f'yolo export model={MODEL}-cls.pt format=torchscript')
|
|
||||||
|
|
||||||
|
|
||||||
def test_export_classify_pose():
|
|
||||||
run(f'yolo export model={MODEL}-pose.pt format=torchscript')
|
|
||||||
|
|
||||||
|
|
||||||
def test_export_detect_edgetpu(enabled=False):
|
|
||||||
if enabled and LINUX:
|
|
||||||
run(f'yolo export model={MODEL}.pt format=edgetpu')
|
|
||||||
|
|||||||
@ -1,6 +1,6 @@
|
|||||||
# Ultralytics YOLO 🚀, AGPL-3.0 license
|
# Ultralytics YOLO 🚀, AGPL-3.0 license
|
||||||
|
|
||||||
__version__ = '8.0.104'
|
__version__ = '8.0.105'
|
||||||
|
|
||||||
from ultralytics.hub import start
|
from ultralytics.hub import start
|
||||||
from ultralytics.vit.rtdetr import RTDETR
|
from ultralytics.vit.rtdetr import RTDETR
|
||||||
|
|||||||
@ -789,13 +789,20 @@ def classify_transforms(size=224, mean=(0.0, 0.0, 0.0), std=(1.0, 1.0, 1.0)): #
|
|||||||
return T.Compose([CenterCrop(size), ToTensor()])
|
return T.Compose([CenterCrop(size), ToTensor()])
|
||||||
|
|
||||||
|
|
||||||
|
def hsv2colorjitter(h, s, v):
|
||||||
|
"""Map HSV (hue, saturation, value) jitter into ColorJitter values (brightness, contrast, saturation, hue)"""
|
||||||
|
return v, v, s, h
|
||||||
|
|
||||||
|
|
||||||
def classify_albumentations(
|
def classify_albumentations(
|
||||||
augment=True,
|
augment=True,
|
||||||
size=224,
|
size=224,
|
||||||
scale=(0.08, 1.0),
|
scale=(0.08, 1.0),
|
||||||
hflip=0.5,
|
hflip=0.5,
|
||||||
vflip=0.0,
|
vflip=0.0,
|
||||||
jitter=0.4,
|
hsv_h=0.015, # image HSV-Hue augmentation (fraction)
|
||||||
|
hsv_s=0.7, # image HSV-Saturation augmentation (fraction)
|
||||||
|
hsv_v=0.4, # image HSV-Value augmentation (fraction)
|
||||||
mean=(0.0, 0.0, 0.0), # IMAGENET_MEAN
|
mean=(0.0, 0.0, 0.0), # IMAGENET_MEAN
|
||||||
std=(1.0, 1.0, 1.0), # IMAGENET_STD
|
std=(1.0, 1.0, 1.0), # IMAGENET_STD
|
||||||
auto_aug=False,
|
auto_aug=False,
|
||||||
@ -810,16 +817,15 @@ def classify_albumentations(
|
|||||||
if augment: # Resize and crop
|
if augment: # Resize and crop
|
||||||
T = [A.RandomResizedCrop(height=size, width=size, scale=scale)]
|
T = [A.RandomResizedCrop(height=size, width=size, scale=scale)]
|
||||||
if auto_aug:
|
if auto_aug:
|
||||||
# TODO: implement AugMix, AutoAug & RandAug in albumentation
|
# TODO: implement AugMix, AutoAug & RandAug in albumentations
|
||||||
LOGGER.info(f'{prefix}auto augmentations are currently not supported')
|
LOGGER.info(f'{prefix}auto augmentations are currently not supported')
|
||||||
else:
|
else:
|
||||||
if hflip > 0:
|
if hflip > 0:
|
||||||
T += [A.HorizontalFlip(p=hflip)]
|
T += [A.HorizontalFlip(p=hflip)]
|
||||||
if vflip > 0:
|
if vflip > 0:
|
||||||
T += [A.VerticalFlip(p=vflip)]
|
T += [A.VerticalFlip(p=vflip)]
|
||||||
if jitter > 0:
|
if any((hsv_h, hsv_s, hsv_v)):
|
||||||
jitter = float(jitter)
|
T += [A.ColorJitter(*hsv2colorjitter(hsv_h, hsv_s, hsv_v))] # brightness, contrast, saturation, hue
|
||||||
T += [A.ColorJitter(jitter, jitter, jitter, 0)] # brightness, contrast, saturation, 0 hue
|
|
||||||
else: # Use fixed crop for eval set (reproducibility)
|
else: # Use fixed crop for eval set (reproducibility)
|
||||||
T = [A.SmallestMaxSize(max_size=size), A.CenterCrop(height=size, width=size)]
|
T = [A.SmallestMaxSize(max_size=size), A.CenterCrop(height=size, width=size)]
|
||||||
T += [A.Normalize(mean=mean, std=std), ToTensorV2()] # Normalize and convert to Tensor
|
T += [A.Normalize(mean=mean, std=std), ToTensorV2()] # Normalize and convert to Tensor
|
||||||
|
|||||||
@ -202,21 +202,48 @@ class YOLODataset(BaseDataset):
|
|||||||
# Classification dataloaders -------------------------------------------------------------------------------------------
|
# Classification dataloaders -------------------------------------------------------------------------------------------
|
||||||
class ClassificationDataset(torchvision.datasets.ImageFolder):
|
class ClassificationDataset(torchvision.datasets.ImageFolder):
|
||||||
"""
|
"""
|
||||||
YOLOv5 Classification Dataset.
|
YOLO Classification Dataset.
|
||||||
Arguments
|
|
||||||
root: Dataset path
|
Args:
|
||||||
transform: torchvision transforms, used by default
|
root (str): Dataset path.
|
||||||
album_transform: Albumentations transforms, used if installed
|
transform (callable, optional): torchvision transforms, used by default.
|
||||||
|
album_transform (callable, optional): Albumentations transforms, used if installed.
|
||||||
|
|
||||||
|
Attributes:
|
||||||
|
cache_ram (bool): True if images should be cached in RAM, False otherwise.
|
||||||
|
cache_disk (bool): True if images should be cached on disk, False otherwise.
|
||||||
|
samples (list): List of samples containing file, index, npy, and im.
|
||||||
|
torch_transforms (callable): torchvision transforms applied to the dataset.
|
||||||
|
album_transforms (callable, optional): Albumentations transforms applied to the dataset if augment is True.
|
||||||
"""
|
"""
|
||||||
|
|
||||||
def __init__(self, root, augment=False, imgsz=224, cache=False):
|
def __init__(self, root, args, augment=False, cache=False):
|
||||||
"""Initialize YOLO object with root, image size, augmentations, and cache settings"""
|
"""
|
||||||
|
Initialize YOLO object with root, image size, augmentations, and cache settings.
|
||||||
|
|
||||||
|
Args:
|
||||||
|
root (str): Dataset path.
|
||||||
|
args (Namespace): Argument parser containing dataset related settings.
|
||||||
|
augment (bool, optional): True if dataset should be augmented, False otherwise. Defaults to False.
|
||||||
|
cache (Union[bool, str], optional): Cache setting, can be True, False, 'ram' or 'disk'. Defaults to False.
|
||||||
|
"""
|
||||||
super().__init__(root=root)
|
super().__init__(root=root)
|
||||||
self.torch_transforms = classify_transforms(imgsz)
|
|
||||||
self.album_transforms = classify_albumentations(augment, imgsz) if augment else None
|
|
||||||
self.cache_ram = cache is True or cache == 'ram'
|
self.cache_ram = cache is True or cache == 'ram'
|
||||||
self.cache_disk = cache == 'disk'
|
self.cache_disk = cache == 'disk'
|
||||||
self.samples = [list(x) + [Path(x[0]).with_suffix('.npy'), None] for x in self.samples] # file, index, npy, im
|
self.samples = [list(x) + [Path(x[0]).with_suffix('.npy'), None] for x in self.samples] # file, index, npy, im
|
||||||
|
self.torch_transforms = classify_transforms(args.imgsz)
|
||||||
|
self.album_transforms = classify_albumentations(
|
||||||
|
augment=augment,
|
||||||
|
size=args.imgsz,
|
||||||
|
scale=(1.0 - args.scale, 1.0), # (0.08, 1.0)
|
||||||
|
hflip=args.fliplr,
|
||||||
|
vflip=args.flipud,
|
||||||
|
hsv_h=args.hsv_h, # HSV-Hue augmentation (fraction)
|
||||||
|
hsv_s=args.hsv_s, # HSV-Saturation augmentation (fraction)
|
||||||
|
hsv_v=args.hsv_v, # HSV-Value augmentation (fraction)
|
||||||
|
mean=(0.0, 0.0, 0.0), # IMAGENET_MEAN
|
||||||
|
std=(1.0, 1.0, 1.0), # IMAGENET_STD
|
||||||
|
auto_aug=False) if augment else None
|
||||||
|
|
||||||
def __getitem__(self, i):
|
def __getitem__(self, i):
|
||||||
"""Returns subset of data and targets corresponding to given indices."""
|
"""Returns subset of data and targets corresponding to given indices."""
|
||||||
|
|||||||
@ -85,6 +85,7 @@ class BaseTrainer:
|
|||||||
self.validator = None
|
self.validator = None
|
||||||
self.model = None
|
self.model = None
|
||||||
self.metrics = None
|
self.metrics = None
|
||||||
|
self.plots = {}
|
||||||
init_seeds(self.args.seed + 1 + RANK, deterministic=self.args.deterministic)
|
init_seeds(self.args.seed + 1 + RANK, deterministic=self.args.deterministic)
|
||||||
|
|
||||||
# Dirs
|
# Dirs
|
||||||
@ -537,6 +538,10 @@ class BaseTrainer:
|
|||||||
"""Plot and display metrics visually."""
|
"""Plot and display metrics visually."""
|
||||||
pass
|
pass
|
||||||
|
|
||||||
|
def on_plot(self, name, data=None):
|
||||||
|
"""Registers plots (e.g. to be consumed in callbacks)"""
|
||||||
|
self.plots[name] = {'data': data, 'timestamp': time.time()}
|
||||||
|
|
||||||
def final_eval(self):
|
def final_eval(self):
|
||||||
"""Performs final evaluation and validation for object detection YOLO model."""
|
"""Performs final evaluation and validation for object detection YOLO model."""
|
||||||
for f in self.last, self.best:
|
for f in self.last, self.best:
|
||||||
|
|||||||
@ -19,6 +19,7 @@ Usage - formats:
|
|||||||
yolov8n_paddle_model # PaddlePaddle
|
yolov8n_paddle_model # PaddlePaddle
|
||||||
"""
|
"""
|
||||||
import json
|
import json
|
||||||
|
import time
|
||||||
from pathlib import Path
|
from pathlib import Path
|
||||||
|
|
||||||
import torch
|
import torch
|
||||||
@ -84,6 +85,7 @@ class BaseValidator:
|
|||||||
if self.args.conf is None:
|
if self.args.conf is None:
|
||||||
self.args.conf = 0.001 # default conf=0.001
|
self.args.conf = 0.001 # default conf=0.001
|
||||||
|
|
||||||
|
self.plots = {}
|
||||||
self.callbacks = _callbacks or callbacks.get_default_callbacks()
|
self.callbacks = _callbacks or callbacks.get_default_callbacks()
|
||||||
|
|
||||||
@smart_inference_mode()
|
@smart_inference_mode()
|
||||||
@ -252,6 +254,10 @@ class BaseValidator:
|
|||||||
"""Returns the metric keys used in YOLO training/validation."""
|
"""Returns the metric keys used in YOLO training/validation."""
|
||||||
return []
|
return []
|
||||||
|
|
||||||
|
def on_plot(self, name, data=None):
|
||||||
|
"""Registers plots (e.g. to be consumed in callbacks)"""
|
||||||
|
self.plots[name] = {'data': data, 'timestamp': time.time()}
|
||||||
|
|
||||||
# TODO: may need to put these following functions into callback
|
# TODO: may need to put these following functions into callback
|
||||||
def plot_val_samples(self, batch, ni):
|
def plot_val_samples(self, batch, ni):
|
||||||
"""Plots validation samples during training."""
|
"""Plots validation samples during training."""
|
||||||
|
|||||||
@ -300,7 +300,7 @@ class ConfusionMatrix:
|
|||||||
|
|
||||||
@TryExcept('WARNING ⚠️ ConfusionMatrix plot failure')
|
@TryExcept('WARNING ⚠️ ConfusionMatrix plot failure')
|
||||||
@plt_settings()
|
@plt_settings()
|
||||||
def plot(self, normalize=True, save_dir='', names=()):
|
def plot(self, normalize=True, save_dir='', names=(), on_plot=None):
|
||||||
"""
|
"""
|
||||||
Plot the confusion matrix using seaborn and save it to a file.
|
Plot the confusion matrix using seaborn and save it to a file.
|
||||||
|
|
||||||
@ -308,6 +308,7 @@ class ConfusionMatrix:
|
|||||||
normalize (bool): Whether to normalize the confusion matrix.
|
normalize (bool): Whether to normalize the confusion matrix.
|
||||||
save_dir (str): Directory where the plot will be saved.
|
save_dir (str): Directory where the plot will be saved.
|
||||||
names (tuple): Names of classes, used as labels on the plot.
|
names (tuple): Names of classes, used as labels on the plot.
|
||||||
|
on_plot (func): An optional callback to pass plots path and data when they are rendered.
|
||||||
"""
|
"""
|
||||||
import seaborn as sn
|
import seaborn as sn
|
||||||
|
|
||||||
@ -336,8 +337,11 @@ class ConfusionMatrix:
|
|||||||
ax.set_xlabel('True')
|
ax.set_xlabel('True')
|
||||||
ax.set_ylabel('Predicted')
|
ax.set_ylabel('Predicted')
|
||||||
ax.set_title(title)
|
ax.set_title(title)
|
||||||
fig.savefig(Path(save_dir) / f'{title.lower().replace(" ", "_")}.png', dpi=250)
|
plot_fname = Path(save_dir) / f'{title.lower().replace(" ", "_")}.png'
|
||||||
|
fig.savefig(plot_fname, dpi=250)
|
||||||
plt.close(fig)
|
plt.close(fig)
|
||||||
|
if on_plot:
|
||||||
|
on_plot(plot_fname)
|
||||||
|
|
||||||
def print(self):
|
def print(self):
|
||||||
"""
|
"""
|
||||||
@ -356,7 +360,7 @@ def smooth(y, f=0.05):
|
|||||||
|
|
||||||
|
|
||||||
@plt_settings()
|
@plt_settings()
|
||||||
def plot_pr_curve(px, py, ap, save_dir=Path('pr_curve.png'), names=()):
|
def plot_pr_curve(px, py, ap, save_dir=Path('pr_curve.png'), names=(), on_plot=None):
|
||||||
"""Plots a precision-recall curve."""
|
"""Plots a precision-recall curve."""
|
||||||
fig, ax = plt.subplots(1, 1, figsize=(9, 6), tight_layout=True)
|
fig, ax = plt.subplots(1, 1, figsize=(9, 6), tight_layout=True)
|
||||||
py = np.stack(py, axis=1)
|
py = np.stack(py, axis=1)
|
||||||
@ -376,10 +380,12 @@ def plot_pr_curve(px, py, ap, save_dir=Path('pr_curve.png'), names=()):
|
|||||||
ax.set_title('Precision-Recall Curve')
|
ax.set_title('Precision-Recall Curve')
|
||||||
fig.savefig(save_dir, dpi=250)
|
fig.savefig(save_dir, dpi=250)
|
||||||
plt.close(fig)
|
plt.close(fig)
|
||||||
|
if on_plot:
|
||||||
|
on_plot(save_dir)
|
||||||
|
|
||||||
|
|
||||||
@plt_settings()
|
@plt_settings()
|
||||||
def plot_mc_curve(px, py, save_dir=Path('mc_curve.png'), names=(), xlabel='Confidence', ylabel='Metric'):
|
def plot_mc_curve(px, py, save_dir=Path('mc_curve.png'), names=(), xlabel='Confidence', ylabel='Metric', on_plot=None):
|
||||||
"""Plots a metric-confidence curve."""
|
"""Plots a metric-confidence curve."""
|
||||||
fig, ax = plt.subplots(1, 1, figsize=(9, 6), tight_layout=True)
|
fig, ax = plt.subplots(1, 1, figsize=(9, 6), tight_layout=True)
|
||||||
|
|
||||||
@ -399,6 +405,8 @@ def plot_mc_curve(px, py, save_dir=Path('mc_curve.png'), names=(), xlabel='Confi
|
|||||||
ax.set_title(f'{ylabel}-Confidence Curve')
|
ax.set_title(f'{ylabel}-Confidence Curve')
|
||||||
fig.savefig(save_dir, dpi=250)
|
fig.savefig(save_dir, dpi=250)
|
||||||
plt.close(fig)
|
plt.close(fig)
|
||||||
|
if on_plot:
|
||||||
|
on_plot(save_dir)
|
||||||
|
|
||||||
|
|
||||||
def compute_ap(recall, precision):
|
def compute_ap(recall, precision):
|
||||||
@ -434,7 +442,16 @@ def compute_ap(recall, precision):
|
|||||||
return ap, mpre, mrec
|
return ap, mpre, mrec
|
||||||
|
|
||||||
|
|
||||||
def ap_per_class(tp, conf, pred_cls, target_cls, plot=False, save_dir=Path(), names=(), eps=1e-16, prefix=''):
|
def ap_per_class(tp,
|
||||||
|
conf,
|
||||||
|
pred_cls,
|
||||||
|
target_cls,
|
||||||
|
plot=False,
|
||||||
|
on_plot=None,
|
||||||
|
save_dir=Path(),
|
||||||
|
names=(),
|
||||||
|
eps=1e-16,
|
||||||
|
prefix=''):
|
||||||
"""
|
"""
|
||||||
Computes the average precision per class for object detection evaluation.
|
Computes the average precision per class for object detection evaluation.
|
||||||
|
|
||||||
@ -444,6 +461,7 @@ def ap_per_class(tp, conf, pred_cls, target_cls, plot=False, save_dir=Path(), na
|
|||||||
pred_cls (np.ndarray): Array of predicted classes of the detections.
|
pred_cls (np.ndarray): Array of predicted classes of the detections.
|
||||||
target_cls (np.ndarray): Array of true classes of the detections.
|
target_cls (np.ndarray): Array of true classes of the detections.
|
||||||
plot (bool, optional): Whether to plot PR curves or not. Defaults to False.
|
plot (bool, optional): Whether to plot PR curves or not. Defaults to False.
|
||||||
|
on_plot (func, optional): A callback to pass plots path and data when they are rendered. Defaults to None.
|
||||||
save_dir (Path, optional): Directory to save the PR curves. Defaults to an empty path.
|
save_dir (Path, optional): Directory to save the PR curves. Defaults to an empty path.
|
||||||
names (tuple, optional): Tuple of class names to plot PR curves. Defaults to an empty tuple.
|
names (tuple, optional): Tuple of class names to plot PR curves. Defaults to an empty tuple.
|
||||||
eps (float, optional): A small value to avoid division by zero. Defaults to 1e-16.
|
eps (float, optional): A small value to avoid division by zero. Defaults to 1e-16.
|
||||||
@ -502,10 +520,10 @@ def ap_per_class(tp, conf, pred_cls, target_cls, plot=False, save_dir=Path(), na
|
|||||||
names = [v for k, v in names.items() if k in unique_classes] # list: only classes that have data
|
names = [v for k, v in names.items() if k in unique_classes] # list: only classes that have data
|
||||||
names = dict(enumerate(names)) # to dict
|
names = dict(enumerate(names)) # to dict
|
||||||
if plot:
|
if plot:
|
||||||
plot_pr_curve(px, py, ap, save_dir / f'{prefix}PR_curve.png', names)
|
plot_pr_curve(px, py, ap, save_dir / f'{prefix}PR_curve.png', names, on_plot=on_plot)
|
||||||
plot_mc_curve(px, f1, save_dir / f'{prefix}F1_curve.png', names, ylabel='F1')
|
plot_mc_curve(px, f1, save_dir / f'{prefix}F1_curve.png', names, ylabel='F1', on_plot=on_plot)
|
||||||
plot_mc_curve(px, p, save_dir / f'{prefix}P_curve.png', names, ylabel='Precision')
|
plot_mc_curve(px, p, save_dir / f'{prefix}P_curve.png', names, ylabel='Precision', on_plot=on_plot)
|
||||||
plot_mc_curve(px, r, save_dir / f'{prefix}R_curve.png', names, ylabel='Recall')
|
plot_mc_curve(px, r, save_dir / f'{prefix}R_curve.png', names, ylabel='Recall', on_plot=on_plot)
|
||||||
|
|
||||||
i = smooth(f1.mean(0), 0.1).argmax() # max F1 index
|
i = smooth(f1.mean(0), 0.1).argmax() # max F1 index
|
||||||
p, r, f1 = p[:, i], r[:, i], f1[:, i]
|
p, r, f1 = p[:, i], r[:, i], f1[:, i]
|
||||||
@ -657,11 +675,13 @@ class DetMetrics(SimpleClass):
|
|||||||
Args:
|
Args:
|
||||||
save_dir (Path): A path to the directory where the output plots will be saved. Defaults to current directory.
|
save_dir (Path): A path to the directory where the output plots will be saved. Defaults to current directory.
|
||||||
plot (bool): A flag that indicates whether to plot precision-recall curves for each class. Defaults to False.
|
plot (bool): A flag that indicates whether to plot precision-recall curves for each class. Defaults to False.
|
||||||
|
on_plot (func): An optional callback to pass plots path and data when they are rendered. Defaults to None.
|
||||||
names (tuple of str): A tuple of strings that represents the names of the classes. Defaults to an empty tuple.
|
names (tuple of str): A tuple of strings that represents the names of the classes. Defaults to an empty tuple.
|
||||||
|
|
||||||
Attributes:
|
Attributes:
|
||||||
save_dir (Path): A path to the directory where the output plots will be saved.
|
save_dir (Path): A path to the directory where the output plots will be saved.
|
||||||
plot (bool): A flag that indicates whether to plot the precision-recall curves for each class.
|
plot (bool): A flag that indicates whether to plot the precision-recall curves for each class.
|
||||||
|
on_plot (func): An optional callback to pass plots path and data when they are rendered.
|
||||||
names (tuple of str): A tuple of strings that represents the names of the classes.
|
names (tuple of str): A tuple of strings that represents the names of the classes.
|
||||||
box (Metric): An instance of the Metric class for storing the results of the detection metrics.
|
box (Metric): An instance of the Metric class for storing the results of the detection metrics.
|
||||||
speed (dict): A dictionary for storing the execution time of different parts of the detection process.
|
speed (dict): A dictionary for storing the execution time of different parts of the detection process.
|
||||||
@ -677,9 +697,10 @@ class DetMetrics(SimpleClass):
|
|||||||
results_dict: Returns a dictionary that maps detection metric keys to their computed values.
|
results_dict: Returns a dictionary that maps detection metric keys to their computed values.
|
||||||
"""
|
"""
|
||||||
|
|
||||||
def __init__(self, save_dir=Path('.'), plot=False, names=()) -> None:
|
def __init__(self, save_dir=Path('.'), plot=False, on_plot=None, names=()) -> None:
|
||||||
self.save_dir = save_dir
|
self.save_dir = save_dir
|
||||||
self.plot = plot
|
self.plot = plot
|
||||||
|
self.on_plot = on_plot
|
||||||
self.names = names
|
self.names = names
|
||||||
self.box = Metric()
|
self.box = Metric()
|
||||||
self.speed = {'preprocess': 0.0, 'inference': 0.0, 'loss': 0.0, 'postprocess': 0.0}
|
self.speed = {'preprocess': 0.0, 'inference': 0.0, 'loss': 0.0, 'postprocess': 0.0}
|
||||||
@ -732,11 +753,13 @@ class SegmentMetrics(SimpleClass):
|
|||||||
Args:
|
Args:
|
||||||
save_dir (Path): Path to the directory where the output plots should be saved. Default is the current directory.
|
save_dir (Path): Path to the directory where the output plots should be saved. Default is the current directory.
|
||||||
plot (bool): Whether to save the detection and segmentation plots. Default is False.
|
plot (bool): Whether to save the detection and segmentation plots. Default is False.
|
||||||
|
on_plot (func): An optional callback to pass plots path and data when they are rendered. Defaults to None.
|
||||||
names (list): List of class names. Default is an empty list.
|
names (list): List of class names. Default is an empty list.
|
||||||
|
|
||||||
Attributes:
|
Attributes:
|
||||||
save_dir (Path): Path to the directory where the output plots should be saved.
|
save_dir (Path): Path to the directory where the output plots should be saved.
|
||||||
plot (bool): Whether to save the detection and segmentation plots.
|
plot (bool): Whether to save the detection and segmentation plots.
|
||||||
|
on_plot (func): An optional callback to pass plots path and data when they are rendered.
|
||||||
names (list): List of class names.
|
names (list): List of class names.
|
||||||
box (Metric): An instance of the Metric class to calculate box detection metrics.
|
box (Metric): An instance of the Metric class to calculate box detection metrics.
|
||||||
seg (Metric): An instance of the Metric class to calculate mask segmentation metrics.
|
seg (Metric): An instance of the Metric class to calculate mask segmentation metrics.
|
||||||
@ -752,9 +775,10 @@ class SegmentMetrics(SimpleClass):
|
|||||||
results_dict: Returns the dictionary containing all the detection and segmentation metrics and fitness score.
|
results_dict: Returns the dictionary containing all the detection and segmentation metrics and fitness score.
|
||||||
"""
|
"""
|
||||||
|
|
||||||
def __init__(self, save_dir=Path('.'), plot=False, names=()) -> None:
|
def __init__(self, save_dir=Path('.'), plot=False, on_plot=None, names=()) -> None:
|
||||||
self.save_dir = save_dir
|
self.save_dir = save_dir
|
||||||
self.plot = plot
|
self.plot = plot
|
||||||
|
self.on_plot = on_plot
|
||||||
self.names = names
|
self.names = names
|
||||||
self.box = Metric()
|
self.box = Metric()
|
||||||
self.seg = Metric()
|
self.seg = Metric()
|
||||||
@ -777,6 +801,7 @@ class SegmentMetrics(SimpleClass):
|
|||||||
pred_cls,
|
pred_cls,
|
||||||
target_cls,
|
target_cls,
|
||||||
plot=self.plot,
|
plot=self.plot,
|
||||||
|
on_plot=self.on_plot,
|
||||||
save_dir=self.save_dir,
|
save_dir=self.save_dir,
|
||||||
names=self.names,
|
names=self.names,
|
||||||
prefix='Mask')[2:]
|
prefix='Mask')[2:]
|
||||||
@ -787,6 +812,7 @@ class SegmentMetrics(SimpleClass):
|
|||||||
pred_cls,
|
pred_cls,
|
||||||
target_cls,
|
target_cls,
|
||||||
plot=self.plot,
|
plot=self.plot,
|
||||||
|
on_plot=self.on_plot,
|
||||||
save_dir=self.save_dir,
|
save_dir=self.save_dir,
|
||||||
names=self.names,
|
names=self.names,
|
||||||
prefix='Box')[2:]
|
prefix='Box')[2:]
|
||||||
@ -836,11 +862,13 @@ class PoseMetrics(SegmentMetrics):
|
|||||||
Args:
|
Args:
|
||||||
save_dir (Path): Path to the directory where the output plots should be saved. Default is the current directory.
|
save_dir (Path): Path to the directory where the output plots should be saved. Default is the current directory.
|
||||||
plot (bool): Whether to save the detection and segmentation plots. Default is False.
|
plot (bool): Whether to save the detection and segmentation plots. Default is False.
|
||||||
|
on_plot (func): An optional callback to pass plots path and data when they are rendered. Defaults to None.
|
||||||
names (list): List of class names. Default is an empty list.
|
names (list): List of class names. Default is an empty list.
|
||||||
|
|
||||||
Attributes:
|
Attributes:
|
||||||
save_dir (Path): Path to the directory where the output plots should be saved.
|
save_dir (Path): Path to the directory where the output plots should be saved.
|
||||||
plot (bool): Whether to save the detection and segmentation plots.
|
plot (bool): Whether to save the detection and segmentation plots.
|
||||||
|
on_plot (func): An optional callback to pass plots path and data when they are rendered.
|
||||||
names (list): List of class names.
|
names (list): List of class names.
|
||||||
box (Metric): An instance of the Metric class to calculate box detection metrics.
|
box (Metric): An instance of the Metric class to calculate box detection metrics.
|
||||||
pose (Metric): An instance of the Metric class to calculate mask segmentation metrics.
|
pose (Metric): An instance of the Metric class to calculate mask segmentation metrics.
|
||||||
@ -856,10 +884,11 @@ class PoseMetrics(SegmentMetrics):
|
|||||||
results_dict: Returns the dictionary containing all the detection and segmentation metrics and fitness score.
|
results_dict: Returns the dictionary containing all the detection and segmentation metrics and fitness score.
|
||||||
"""
|
"""
|
||||||
|
|
||||||
def __init__(self, save_dir=Path('.'), plot=False, names=()) -> None:
|
def __init__(self, save_dir=Path('.'), plot=False, on_plot=None, names=()) -> None:
|
||||||
super().__init__(save_dir, plot, names)
|
super().__init__(save_dir, plot, names)
|
||||||
self.save_dir = save_dir
|
self.save_dir = save_dir
|
||||||
self.plot = plot
|
self.plot = plot
|
||||||
|
self.on_plot = on_plot
|
||||||
self.names = names
|
self.names = names
|
||||||
self.box = Metric()
|
self.box = Metric()
|
||||||
self.pose = Metric()
|
self.pose = Metric()
|
||||||
@ -887,6 +916,7 @@ class PoseMetrics(SegmentMetrics):
|
|||||||
pred_cls,
|
pred_cls,
|
||||||
target_cls,
|
target_cls,
|
||||||
plot=self.plot,
|
plot=self.plot,
|
||||||
|
on_plot=self.on_plot,
|
||||||
save_dir=self.save_dir,
|
save_dir=self.save_dir,
|
||||||
names=self.names,
|
names=self.names,
|
||||||
prefix='Pose')[2:]
|
prefix='Pose')[2:]
|
||||||
@ -897,6 +927,7 @@ class PoseMetrics(SegmentMetrics):
|
|||||||
pred_cls,
|
pred_cls,
|
||||||
target_cls,
|
target_cls,
|
||||||
plot=self.plot,
|
plot=self.plot,
|
||||||
|
on_plot=self.on_plot,
|
||||||
save_dir=self.save_dir,
|
save_dir=self.save_dir,
|
||||||
names=self.names,
|
names=self.names,
|
||||||
prefix='Box')[2:]
|
prefix='Box')[2:]
|
||||||
|
|||||||
@ -228,7 +228,7 @@ class Annotator:
|
|||||||
|
|
||||||
@TryExcept() # known issue https://github.com/ultralytics/yolov5/issues/5395
|
@TryExcept() # known issue https://github.com/ultralytics/yolov5/issues/5395
|
||||||
@plt_settings()
|
@plt_settings()
|
||||||
def plot_labels(boxes, cls, names=(), save_dir=Path('')):
|
def plot_labels(boxes, cls, names=(), save_dir=Path(''), on_plot=None):
|
||||||
"""Save and plot image with no axis or spines."""
|
"""Save and plot image with no axis or spines."""
|
||||||
import pandas as pd
|
import pandas as pd
|
||||||
import seaborn as sn
|
import seaborn as sn
|
||||||
@ -271,8 +271,11 @@ def plot_labels(boxes, cls, names=(), save_dir=Path('')):
|
|||||||
for s in ['top', 'right', 'left', 'bottom']:
|
for s in ['top', 'right', 'left', 'bottom']:
|
||||||
ax[a].spines[s].set_visible(False)
|
ax[a].spines[s].set_visible(False)
|
||||||
|
|
||||||
plt.savefig(save_dir / 'labels.jpg', dpi=200)
|
fname = save_dir / 'labels.jpg'
|
||||||
|
plt.savefig(fname, dpi=200)
|
||||||
plt.close()
|
plt.close()
|
||||||
|
if on_plot:
|
||||||
|
on_plot(fname)
|
||||||
|
|
||||||
|
|
||||||
def save_one_box(xyxy, im, file=Path('im.jpg'), gain=1.02, pad=10, square=False, BGR=False, save=True):
|
def save_one_box(xyxy, im, file=Path('im.jpg'), gain=1.02, pad=10, square=False, BGR=False, save=True):
|
||||||
@ -301,7 +304,8 @@ def plot_images(images,
|
|||||||
kpts=np.zeros((0, 51), dtype=np.float32),
|
kpts=np.zeros((0, 51), dtype=np.float32),
|
||||||
paths=None,
|
paths=None,
|
||||||
fname='images.jpg',
|
fname='images.jpg',
|
||||||
names=None):
|
names=None,
|
||||||
|
on_plot=None):
|
||||||
# Plot image grid with labels
|
# Plot image grid with labels
|
||||||
if isinstance(images, torch.Tensor):
|
if isinstance(images, torch.Tensor):
|
||||||
images = images.cpu().float().numpy()
|
images = images.cpu().float().numpy()
|
||||||
@ -419,10 +423,12 @@ def plot_images(images,
|
|||||||
im[y:y + h, x:x + w, :][mask] = im[y:y + h, x:x + w, :][mask] * 0.4 + np.array(color) * 0.6
|
im[y:y + h, x:x + w, :][mask] = im[y:y + h, x:x + w, :][mask] * 0.4 + np.array(color) * 0.6
|
||||||
annotator.fromarray(im)
|
annotator.fromarray(im)
|
||||||
annotator.im.save(fname) # save
|
annotator.im.save(fname) # save
|
||||||
|
if on_plot:
|
||||||
|
on_plot(fname)
|
||||||
|
|
||||||
|
|
||||||
@plt_settings()
|
@plt_settings()
|
||||||
def plot_results(file='path/to/results.csv', dir='', segment=False, pose=False, classify=False):
|
def plot_results(file='path/to/results.csv', dir='', segment=False, pose=False, classify=False, on_plot=None):
|
||||||
"""Plot training results.csv. Usage: from utils.plots import *; plot_results('path/to/results.csv')."""
|
"""Plot training results.csv. Usage: from utils.plots import *; plot_results('path/to/results.csv')."""
|
||||||
import pandas as pd
|
import pandas as pd
|
||||||
save_dir = Path(file).parent if file else Path(dir)
|
save_dir = Path(file).parent if file else Path(dir)
|
||||||
@ -456,8 +462,11 @@ def plot_results(file='path/to/results.csv', dir='', segment=False, pose=False,
|
|||||||
except Exception as e:
|
except Exception as e:
|
||||||
LOGGER.warning(f'WARNING: Plotting error for {f}: {e}')
|
LOGGER.warning(f'WARNING: Plotting error for {f}: {e}')
|
||||||
ax[1].legend()
|
ax[1].legend()
|
||||||
fig.savefig(save_dir / 'results.png', dpi=200)
|
fname = save_dir / 'results.png'
|
||||||
|
fig.savefig(fname, dpi=200)
|
||||||
plt.close()
|
plt.close()
|
||||||
|
if on_plot:
|
||||||
|
on_plot(fname)
|
||||||
|
|
||||||
|
|
||||||
def output_to_target(output, max_det=300):
|
def output_to_target(output, max_det=300):
|
||||||
|
|||||||
@ -71,7 +71,7 @@ class ClassificationTrainer(BaseTrainer):
|
|||||||
return # dont return ckpt. Classification doesn't support resume
|
return # dont return ckpt. Classification doesn't support resume
|
||||||
|
|
||||||
def build_dataset(self, img_path, mode='train', batch=None):
|
def build_dataset(self, img_path, mode='train', batch=None):
|
||||||
return ClassificationDataset(root=img_path, imgsz=self.args.imgsz, augment=mode == 'train')
|
return ClassificationDataset(root=img_path, args=self.args, augment=mode == 'train')
|
||||||
|
|
||||||
def get_dataloader(self, dataset_path, batch_size=16, rank=0, mode='train'):
|
def get_dataloader(self, dataset_path, batch_size=16, rank=0, mode='train'):
|
||||||
"""Returns PyTorch DataLoader with transforms to preprocess images for inference."""
|
"""Returns PyTorch DataLoader with transforms to preprocess images for inference."""
|
||||||
@ -126,7 +126,7 @@ class ClassificationTrainer(BaseTrainer):
|
|||||||
|
|
||||||
def plot_metrics(self):
|
def plot_metrics(self):
|
||||||
"""Plots metrics from a CSV file."""
|
"""Plots metrics from a CSV file."""
|
||||||
plot_results(file=self.csv, classify=True) # save results.png
|
plot_results(file=self.csv, classify=True, on_plot=self.on_plot) # save results.png
|
||||||
|
|
||||||
def final_eval(self):
|
def final_eval(self):
|
||||||
"""Evaluate trained model and save validation results."""
|
"""Evaluate trained model and save validation results."""
|
||||||
@ -147,7 +147,8 @@ class ClassificationTrainer(BaseTrainer):
|
|||||||
plot_images(images=batch['img'],
|
plot_images(images=batch['img'],
|
||||||
batch_idx=torch.arange(len(batch['img'])),
|
batch_idx=torch.arange(len(batch['img'])),
|
||||||
cls=batch['cls'].squeeze(-1),
|
cls=batch['cls'].squeeze(-1),
|
||||||
fname=self.save_dir / f'train_batch{ni}.jpg')
|
fname=self.save_dir / f'train_batch{ni}.jpg',
|
||||||
|
on_plot=self.on_plot)
|
||||||
|
|
||||||
|
|
||||||
def train(cfg=DEFAULT_CFG, use_python=False):
|
def train(cfg=DEFAULT_CFG, use_python=False):
|
||||||
|
|||||||
@ -47,7 +47,10 @@ class ClassificationValidator(BaseValidator):
|
|||||||
self.confusion_matrix.process_cls_preds(self.pred, self.targets)
|
self.confusion_matrix.process_cls_preds(self.pred, self.targets)
|
||||||
if self.args.plots:
|
if self.args.plots:
|
||||||
for normalize in True, False:
|
for normalize in True, False:
|
||||||
self.confusion_matrix.plot(save_dir=self.save_dir, names=self.names.values(), normalize=normalize)
|
self.confusion_matrix.plot(save_dir=self.save_dir,
|
||||||
|
names=self.names.values(),
|
||||||
|
normalize=normalize,
|
||||||
|
on_plot=self.on_plot)
|
||||||
self.metrics.speed = self.speed
|
self.metrics.speed = self.speed
|
||||||
self.metrics.confusion_matrix = self.confusion_matrix
|
self.metrics.confusion_matrix = self.confusion_matrix
|
||||||
|
|
||||||
@ -57,7 +60,7 @@ class ClassificationValidator(BaseValidator):
|
|||||||
return self.metrics.results_dict
|
return self.metrics.results_dict
|
||||||
|
|
||||||
def build_dataset(self, img_path):
|
def build_dataset(self, img_path):
|
||||||
dataset = ClassificationDataset(root=img_path, imgsz=self.args.imgsz, augment=False)
|
dataset = ClassificationDataset(root=img_path, args=self.args, augment=False)
|
||||||
return dataset
|
return dataset
|
||||||
|
|
||||||
def get_dataloader(self, dataset_path, batch_size):
|
def get_dataloader(self, dataset_path, batch_size):
|
||||||
@ -76,7 +79,8 @@ class ClassificationValidator(BaseValidator):
|
|||||||
batch_idx=torch.arange(len(batch['img'])),
|
batch_idx=torch.arange(len(batch['img'])),
|
||||||
cls=batch['cls'].squeeze(-1),
|
cls=batch['cls'].squeeze(-1),
|
||||||
fname=self.save_dir / f'val_batch{ni}_labels.jpg',
|
fname=self.save_dir / f'val_batch{ni}_labels.jpg',
|
||||||
names=self.names)
|
names=self.names,
|
||||||
|
on_plot=self.on_plot)
|
||||||
|
|
||||||
def plot_predictions(self, batch, preds, ni):
|
def plot_predictions(self, batch, preds, ni):
|
||||||
"""Plots predicted bounding boxes on input images and saves the result."""
|
"""Plots predicted bounding boxes on input images and saves the result."""
|
||||||
@ -84,7 +88,8 @@ class ClassificationValidator(BaseValidator):
|
|||||||
batch_idx=torch.arange(len(batch['img'])),
|
batch_idx=torch.arange(len(batch['img'])),
|
||||||
cls=torch.argmax(preds, dim=1),
|
cls=torch.argmax(preds, dim=1),
|
||||||
fname=self.save_dir / f'val_batch{ni}_pred.jpg',
|
fname=self.save_dir / f'val_batch{ni}_pred.jpg',
|
||||||
names=self.names) # pred
|
names=self.names,
|
||||||
|
on_plot=self.on_plot) # pred
|
||||||
|
|
||||||
|
|
||||||
def val(cfg=DEFAULT_CFG, use_python=False):
|
def val(cfg=DEFAULT_CFG, use_python=False):
|
||||||
|
|||||||
@ -121,17 +121,18 @@ class DetectionTrainer(BaseTrainer):
|
|||||||
cls=batch['cls'].squeeze(-1),
|
cls=batch['cls'].squeeze(-1),
|
||||||
bboxes=batch['bboxes'],
|
bboxes=batch['bboxes'],
|
||||||
paths=batch['im_file'],
|
paths=batch['im_file'],
|
||||||
fname=self.save_dir / f'train_batch{ni}.jpg')
|
fname=self.save_dir / f'train_batch{ni}.jpg',
|
||||||
|
on_plot=self.on_plot)
|
||||||
|
|
||||||
def plot_metrics(self):
|
def plot_metrics(self):
|
||||||
"""Plots metrics from a CSV file."""
|
"""Plots metrics from a CSV file."""
|
||||||
plot_results(file=self.csv) # save results.png
|
plot_results(file=self.csv, on_plot=self.on_plot) # save results.png
|
||||||
|
|
||||||
def plot_training_labels(self):
|
def plot_training_labels(self):
|
||||||
"""Create a labeled training plot of the YOLO model."""
|
"""Create a labeled training plot of the YOLO model."""
|
||||||
boxes = np.concatenate([lb['bboxes'] for lb in self.train_loader.dataset.labels], 0)
|
boxes = np.concatenate([lb['bboxes'] for lb in self.train_loader.dataset.labels], 0)
|
||||||
cls = np.concatenate([lb['cls'] for lb in self.train_loader.dataset.labels], 0)
|
cls = np.concatenate([lb['cls'] for lb in self.train_loader.dataset.labels], 0)
|
||||||
plot_labels(boxes, cls.squeeze(), names=self.data['names'], save_dir=self.save_dir)
|
plot_labels(boxes, cls.squeeze(), names=self.data['names'], save_dir=self.save_dir, on_plot=self.on_plot)
|
||||||
|
|
||||||
|
|
||||||
# Criterion class for computing training losses
|
# Criterion class for computing training losses
|
||||||
|
|||||||
@ -24,7 +24,7 @@ class DetectionValidator(BaseValidator):
|
|||||||
self.args.task = 'detect'
|
self.args.task = 'detect'
|
||||||
self.is_coco = False
|
self.is_coco = False
|
||||||
self.class_map = None
|
self.class_map = None
|
||||||
self.metrics = DetMetrics(save_dir=self.save_dir)
|
self.metrics = DetMetrics(save_dir=self.save_dir, on_plot=self.on_plot)
|
||||||
self.iouv = torch.linspace(0.5, 0.95, 10) # iou vector for mAP@0.5:0.95
|
self.iouv = torch.linspace(0.5, 0.95, 10) # iou vector for mAP@0.5:0.95
|
||||||
self.niou = self.iouv.numel()
|
self.niou = self.iouv.numel()
|
||||||
|
|
||||||
@ -145,7 +145,10 @@ class DetectionValidator(BaseValidator):
|
|||||||
|
|
||||||
if self.args.plots:
|
if self.args.plots:
|
||||||
for normalize in True, False:
|
for normalize in True, False:
|
||||||
self.confusion_matrix.plot(save_dir=self.save_dir, names=self.names.values(), normalize=normalize)
|
self.confusion_matrix.plot(save_dir=self.save_dir,
|
||||||
|
names=self.names.values(),
|
||||||
|
normalize=normalize,
|
||||||
|
on_plot=self.on_plot)
|
||||||
|
|
||||||
def _process_batch(self, detections, labels):
|
def _process_batch(self, detections, labels):
|
||||||
"""
|
"""
|
||||||
@ -215,7 +218,8 @@ class DetectionValidator(BaseValidator):
|
|||||||
batch['bboxes'],
|
batch['bboxes'],
|
||||||
paths=batch['im_file'],
|
paths=batch['im_file'],
|
||||||
fname=self.save_dir / f'val_batch{ni}_labels.jpg',
|
fname=self.save_dir / f'val_batch{ni}_labels.jpg',
|
||||||
names=self.names)
|
names=self.names,
|
||||||
|
on_plot=self.on_plot)
|
||||||
|
|
||||||
def plot_predictions(self, batch, preds, ni):
|
def plot_predictions(self, batch, preds, ni):
|
||||||
"""Plots predicted bounding boxes on input images and saves the result."""
|
"""Plots predicted bounding boxes on input images and saves the result."""
|
||||||
@ -223,7 +227,8 @@ class DetectionValidator(BaseValidator):
|
|||||||
*output_to_target(preds, max_det=15),
|
*output_to_target(preds, max_det=15),
|
||||||
paths=batch['im_file'],
|
paths=batch['im_file'],
|
||||||
fname=self.save_dir / f'val_batch{ni}_pred.jpg',
|
fname=self.save_dir / f'val_batch{ni}_pred.jpg',
|
||||||
names=self.names) # pred
|
names=self.names,
|
||||||
|
on_plot=self.on_plot) # pred
|
||||||
|
|
||||||
def save_one_txt(self, predn, save_conf, shape, file):
|
def save_one_txt(self, predn, save_conf, shape, file):
|
||||||
"""Save YOLO detections to a txt file in normalized coordinates in a specific format."""
|
"""Save YOLO detections to a txt file in normalized coordinates in a specific format."""
|
||||||
|
|||||||
@ -65,11 +65,12 @@ class PoseTrainer(v8.detect.DetectionTrainer):
|
|||||||
bboxes,
|
bboxes,
|
||||||
kpts=kpts,
|
kpts=kpts,
|
||||||
paths=paths,
|
paths=paths,
|
||||||
fname=self.save_dir / f'train_batch{ni}.jpg')
|
fname=self.save_dir / f'train_batch{ni}.jpg',
|
||||||
|
on_plot=self.on_plot)
|
||||||
|
|
||||||
def plot_metrics(self):
|
def plot_metrics(self):
|
||||||
"""Plots training/val metrics."""
|
"""Plots training/val metrics."""
|
||||||
plot_results(file=self.csv, pose=True) # save results.png
|
plot_results(file=self.csv, pose=True, on_plot=self.on_plot) # save results.png
|
||||||
|
|
||||||
|
|
||||||
# Criterion class for computing training losses
|
# Criterion class for computing training losses
|
||||||
|
|||||||
@ -18,7 +18,7 @@ class PoseValidator(DetectionValidator):
|
|||||||
"""Initialize a 'PoseValidator' object with custom parameters and assigned attributes."""
|
"""Initialize a 'PoseValidator' object with custom parameters and assigned attributes."""
|
||||||
super().__init__(dataloader, save_dir, pbar, args, _callbacks)
|
super().__init__(dataloader, save_dir, pbar, args, _callbacks)
|
||||||
self.args.task = 'pose'
|
self.args.task = 'pose'
|
||||||
self.metrics = PoseMetrics(save_dir=self.save_dir)
|
self.metrics = PoseMetrics(save_dir=self.save_dir, on_plot=self.on_plot)
|
||||||
|
|
||||||
def preprocess(self, batch):
|
def preprocess(self, batch):
|
||||||
"""Preprocesses the batch by converting the 'keypoints' data into a float and moving it to the device."""
|
"""Preprocesses the batch by converting the 'keypoints' data into a float and moving it to the device."""
|
||||||
@ -150,7 +150,8 @@ class PoseValidator(DetectionValidator):
|
|||||||
kpts=batch['keypoints'],
|
kpts=batch['keypoints'],
|
||||||
paths=batch['im_file'],
|
paths=batch['im_file'],
|
||||||
fname=self.save_dir / f'val_batch{ni}_labels.jpg',
|
fname=self.save_dir / f'val_batch{ni}_labels.jpg',
|
||||||
names=self.names)
|
names=self.names,
|
||||||
|
on_plot=self.on_plot)
|
||||||
|
|
||||||
def plot_predictions(self, batch, preds, ni):
|
def plot_predictions(self, batch, preds, ni):
|
||||||
"""Plots predictions for YOLO model."""
|
"""Plots predictions for YOLO model."""
|
||||||
@ -160,7 +161,8 @@ class PoseValidator(DetectionValidator):
|
|||||||
kpts=pred_kpts,
|
kpts=pred_kpts,
|
||||||
paths=batch['im_file'],
|
paths=batch['im_file'],
|
||||||
fname=self.save_dir / f'val_batch{ni}_pred.jpg',
|
fname=self.save_dir / f'val_batch{ni}_pred.jpg',
|
||||||
names=self.names) # pred
|
names=self.names,
|
||||||
|
on_plot=self.on_plot) # pred
|
||||||
|
|
||||||
def pred_to_json(self, predn, filename):
|
def pred_to_json(self, predn, filename):
|
||||||
"""Converts YOLO predictions to COCO JSON format."""
|
"""Converts YOLO predictions to COCO JSON format."""
|
||||||
|
|||||||
@ -45,17 +45,18 @@ class SegmentationTrainer(v8.detect.DetectionTrainer):
|
|||||||
|
|
||||||
def plot_training_samples(self, batch, ni):
|
def plot_training_samples(self, batch, ni):
|
||||||
"""Creates a plot of training sample images with labels and box coordinates."""
|
"""Creates a plot of training sample images with labels and box coordinates."""
|
||||||
images = batch['img']
|
plot_images(batch['img'],
|
||||||
masks = batch['masks']
|
batch['batch_idx'],
|
||||||
cls = batch['cls'].squeeze(-1)
|
batch['cls'].squeeze(-1),
|
||||||
bboxes = batch['bboxes']
|
batch['bboxes'],
|
||||||
paths = batch['im_file']
|
batch['masks'],
|
||||||
batch_idx = batch['batch_idx']
|
paths=batch['im_file'],
|
||||||
plot_images(images, batch_idx, cls, bboxes, masks, paths=paths, fname=self.save_dir / f'train_batch{ni}.jpg')
|
fname=self.save_dir / f'train_batch{ni}.jpg',
|
||||||
|
on_plot=self.on_plot)
|
||||||
|
|
||||||
def plot_metrics(self):
|
def plot_metrics(self):
|
||||||
"""Plots training/val metrics."""
|
"""Plots training/val metrics."""
|
||||||
plot_results(file=self.csv, segment=True) # save results.png
|
plot_results(file=self.csv, segment=True, on_plot=self.on_plot) # save results.png
|
||||||
|
|
||||||
|
|
||||||
# Criterion class for computing training losses
|
# Criterion class for computing training losses
|
||||||
|
|||||||
@ -20,7 +20,7 @@ class SegmentationValidator(DetectionValidator):
|
|||||||
"""Initialize SegmentationValidator and set task to 'segment', metrics to SegmentMetrics."""
|
"""Initialize SegmentationValidator and set task to 'segment', metrics to SegmentMetrics."""
|
||||||
super().__init__(dataloader, save_dir, pbar, args, _callbacks)
|
super().__init__(dataloader, save_dir, pbar, args, _callbacks)
|
||||||
self.args.task = 'segment'
|
self.args.task = 'segment'
|
||||||
self.metrics = SegmentMetrics(save_dir=self.save_dir)
|
self.metrics = SegmentMetrics(save_dir=self.save_dir, on_plot=self.on_plot)
|
||||||
|
|
||||||
def preprocess(self, batch):
|
def preprocess(self, batch):
|
||||||
"""Preprocesses batch by converting masks to float and sending to device."""
|
"""Preprocesses batch by converting masks to float and sending to device."""
|
||||||
@ -174,7 +174,8 @@ class SegmentationValidator(DetectionValidator):
|
|||||||
batch['masks'],
|
batch['masks'],
|
||||||
paths=batch['im_file'],
|
paths=batch['im_file'],
|
||||||
fname=self.save_dir / f'val_batch{ni}_labels.jpg',
|
fname=self.save_dir / f'val_batch{ni}_labels.jpg',
|
||||||
names=self.names)
|
names=self.names,
|
||||||
|
on_plot=self.on_plot)
|
||||||
|
|
||||||
def plot_predictions(self, batch, preds, ni):
|
def plot_predictions(self, batch, preds, ni):
|
||||||
"""Plots batch predictions with masks and bounding boxes."""
|
"""Plots batch predictions with masks and bounding boxes."""
|
||||||
@ -183,7 +184,8 @@ class SegmentationValidator(DetectionValidator):
|
|||||||
torch.cat(self.plot_masks, dim=0) if len(self.plot_masks) else self.plot_masks,
|
torch.cat(self.plot_masks, dim=0) if len(self.plot_masks) else self.plot_masks,
|
||||||
paths=batch['im_file'],
|
paths=batch['im_file'],
|
||||||
fname=self.save_dir / f'val_batch{ni}_pred.jpg',
|
fname=self.save_dir / f'val_batch{ni}_pred.jpg',
|
||||||
names=self.names) # pred
|
names=self.names,
|
||||||
|
on_plot=self.on_plot) # pred
|
||||||
self.plot_masks.clear()
|
self.plot_masks.clear()
|
||||||
|
|
||||||
def pred_to_json(self, predn, filename, pred_masks):
|
def pred_to_json(self, predn, filename, pred_masks):
|
||||||
|
|||||||
Reference in New Issue
Block a user